Keras 3d U‑net For Multiclass Brain Tumor Segmentation (4 Classes)
تفاصيل العمل
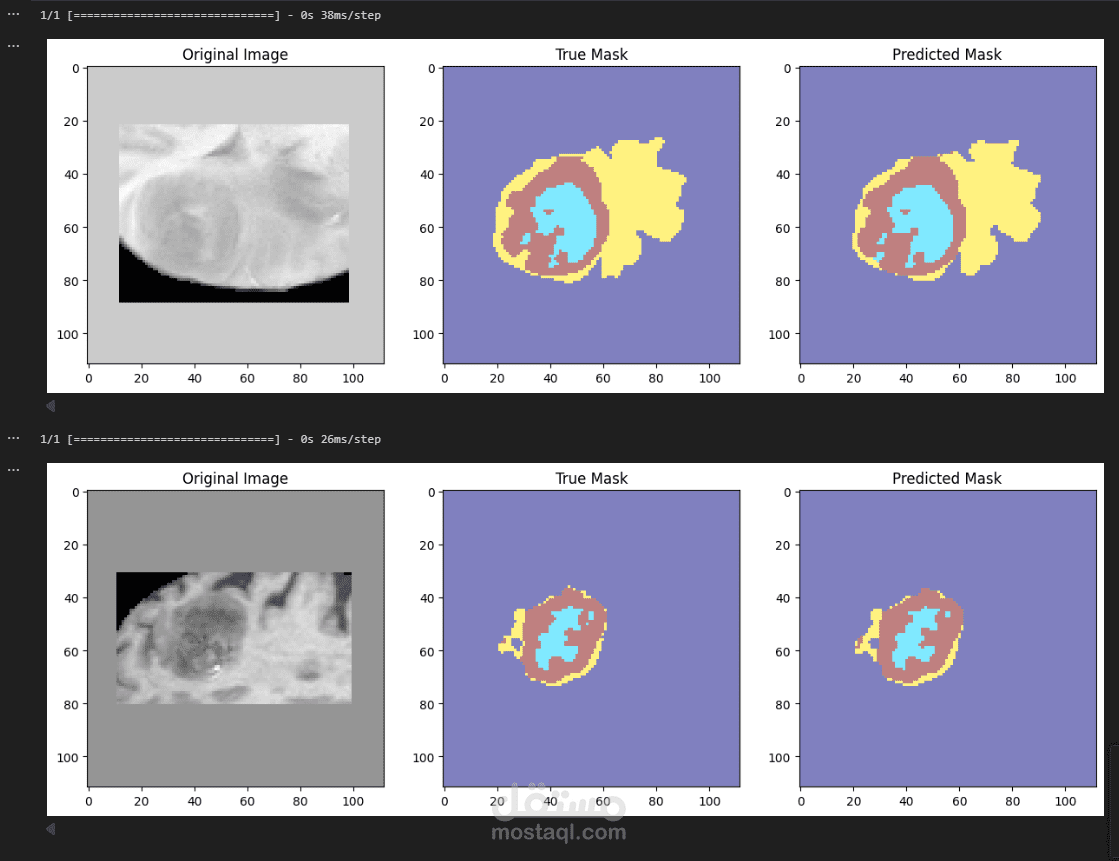
3D Multiclass Brain Tumor Segmentation
This project implements a deep learning pipeline for 3D brain tumor segmentation using multi-modal MRI scans. It is designed to detect and classify different tumor subregions in volumetric brain images.
Key Features
Input: Four MRI modalities per patient (T1, T1ce, T2, FLAIR).
Output: Voxel-wise segmentation map with 4 classes:
0 – Background
1 – Tumor Region A (shown in red)
2 – Tumor Region B (shown in green)
3 – Tumor Region C (shown in blue)
Model: 3D U-Net with encoder–decoder architecture.
Loss Function: Combined Dice + Cross-Entropy for robust multiclass segmentation.
Metrics: Per-class Dice and mean Dice score.
Visualization: 2D slice viewer and interactive 3D tumor rendering (via VTK.js in the Flask frontend).
Deployment: Flask web application where users can upload MRI volumes, run segmentation, and visualize results in both 2D and 3D.
Workflow
Preprocess MRI scans (resampling, normalization, augmentation).
Train 3D U-Net on labeled MRI datasets.
Predict tumor regions and export segmentation masks.
Visualize results interactively in the web app (slice-by-slice and 3D tumor view).
Applications
Medical image analysis & tumor monitoring.
Clinical decision support for neurosurgeons and radiologists.
Research in brain tumor progression and treatment planning.